Granger Processing Stream (GPS)
Our analyses of directed information flow between brain regions (effective connectivity) rely on the Granger Processing Stream or GPS. GPS is a GUI-driven analysis tool for performing Kalman-filter driven Granger Causation Analysis of MR-constrained source space reconstructions of simultaneously acquired MEG and EEG data. Developed by Sir Clive Granger based on the work of Norbert Weiner, Granger identifies patterns of activation consistent with causation based on the observation that causes both precede, and uniquely predict effects. In simple mathematical terms, unique prediction can be quantified by comparing the error terms of predictive models of the activity of one brain region based on prior patterns of all activation of all potentially causal brain regions, with error terms from predictive models of the same region that leave potential causal brain area out. Granger causation is indexed by the log ratio of these two error terms.
GPS acts as a shell program for reconstructing MR data in Freesurfer and reconstructing MEG and EEG data in MNE, widely used tools produced and supported by our colleagues at the Athinoula A. Martinos Center for Biomedical Imaging. We use these tools to create sourcespace reconstructions of brain activity over all cortical surfaces, and then use data-driven techniques to identify a set of brain regions that optimally satisfy the assumptions of Granger causation. GPS then uses a kalmen filter technique to build predictive models for networks that frequently include 30-50 brain regions. Because Kalman filter models are instantaneous, this approach means that we do not have to deal with the non-stationarity of evoked neural patterns (a major problem for implementations of Granger analysis). Moreover, they make it possible to assess evolving patterns of effective connectivity over large networks at millisecond intervals
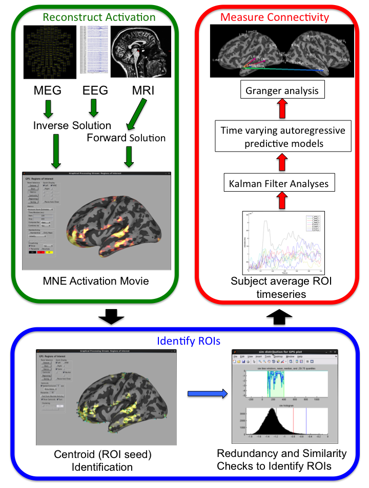
The GPS processing suite provides tools for visualizing global and local patterns of effective connectivity over circle graphs and a variety of cortical representations, as well as performing inferential statistics to compare patterns of effective connectivity between different experimental conditions.
The GPS processing suite is freely available for download at https://www.nmr.mgh.harvard.edu/software/gps, along with a manual, and step-by-step cookbook for performing basic analyses. An overview lecture on GPS can be found at https://cbmm.mit.edu/meg-workshop. Animated visualizations of GPS analyses can be found here